Published On June 23, 2013
DURING THE LATE 1990S, Linda Griffith was working in the laboratory of Joseph Vacanti, a pioneer in the field of tissue engineering, when she had a revelation. Vacanti, Griffith and their colleagues at Massachusetts General Hospital were working to build a transplantable liver. “The challenges were daunting,” says Griffith, now a professor of biological and mechanical engineering at MIT.
Modern tissue engineering—defined by Vacanti and MIT professor Robert Langer in a 1993 paper in Science—applies concepts and techniques from engineering and life sciences to the problem of cultivating human tissue and whole organs to repair or replace damaged parts of the body. Typically, this involves creating a “scaffold” of natural or synthetic materials, seeding it with human stem cells that can differentiate themselves into particular tissue types, and providing the cells with nutrients and a physical environment that encourages them to take on the three-dimensional structures and functions of a particular body part.
The field has had notable successes—there are people walking around today with lab-grown skin, cartilage, bladders and tracheas—but the goal of building a liver, heart or another large, complex organ remains elusive. Sheer mass is a major issue—it’s very difficult to nourish a 3-D population of cells in the lab on the scale of a liver. And even if you succeed in growing such an organ, transferring it from sterile, controlled conditions into the bloody, chaotic environment of a human being presents its own array of problems.
As Griffith struggled with the technical limitations of her work, she thought more and more about the clinical issues her research was addressing—and her engineer’s mind characteristically flipped the problem. Instead of building a replacement liver, she thought, why not use the same underlying techniques for growing cells in three dimensions to create a better model of how a liver behaves? “As a patient, I’d rather have a cure for my disease, or know how to prevent it, than have an organ transplant,” says Griffith. “It’s very exciting to think about growing a liver or a heart. But I realized that, practically, I could have a lot more impact creating organ models.”
Today, Griffith is a leader in a burgeoning subfield of tissue engineering that does just that: creates microversions of living human organs. These “organs on a chip”—so called for their resemblance to silicon computer microchips—look nothing like the body parts they’re meant to replicate. Generally no bigger than a computer thumb drive and made of clear plastic, the devices contain tiny chambers embedded with populations of living cells and connected by microfluidic channels that control the flow of nutrients and oxygen through the system, mimicking the essential structures and functions of organs such as the lung or liver. Such abilities are proving remarkably useful, with the potential to do everything from reducing dependence on animal models and speeding drug development to demonstrating disease processes—and even, in a neat twist, helping researchers engineer entire implantable organs.
GRIFFITH WAS HARDLY THE ONLY PERSON to make the intellectual leap from engineering organs to creating better models of them. Also during the late 1990s, Cornell University researchers Michael Shuler and Gregory Baxter, for example, described an “animal on a chip” that used living cells embedded in a silicon chip. Since then, drawing on micromanufacturing technologies borrowed from the computer industry, researchers have tinkered with a variety of human-tissue-and-chip constructions, developing chips for the lung, heart, liver, kidney, bone marrow, brain and gut. And with a flurry of funding in 2012 from the National Institutes of Health and DARPA, the U.S. military’s research arm, the field has gained momentum and a higher profile.
The growing interest stems largely from a sense of frustration among scientists and pharmaceutical companies with existing methods of testing how well new drugs work and whether or not they may be toxic. Drug development typically begins with a screening that tests the effects of promising compounds on two-dimensional cell cultures. Drug candidates that pass this stage are tested for toxicity in animals before advancing to human trials. Yet, according to the NIH, 30% of compounds that get past those early hurdles still turn out to be toxic to people. Such dead ends are a major contributor to rising drug development costs, estimated to be at least $4 billion for a single new drug from a major manufacturer.
“The predictions animal models make are often wrong,” says Don Ingber, who heads the human-on-a-chip project at Harvard’s Wyss Institute. “It’s killing the pharmaceutical industry. The opposite of Moore’s Law is Eroom’s Law [Moore spelled backward], which says that the number of medicines invented halves each year, while the prices seem to go up continually.” Ingber and others believe organs on chips can help identify ineffective or unsafe drugs much earlier in the development process while at the same time reducing if not eliminating the need for animal testing. That could also accelerate the arrival of new drugs—for example, to treat an emerging disease that threatens public health—which helps explain DARPA’s interest in organ-on-a-chip research.
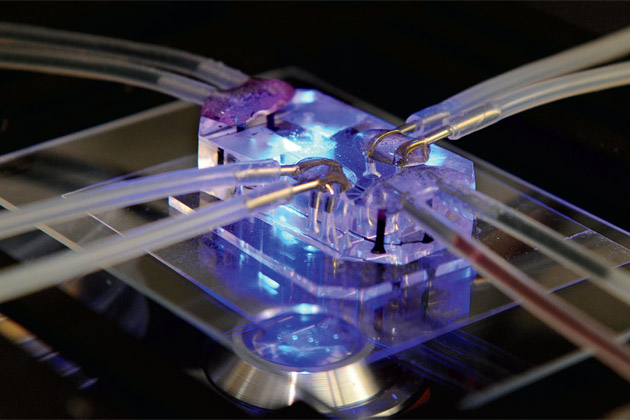
A TINY VERSION OF AN ORGAN has many advantages over a full-size model. To start with, there’s cost and convenience. Human cells are expensive. So using as few of them as possible to create models of human tissue and organs saves money. Small models of organs are also easier to interrogate and understand than large ones. “If we go life-size, we have the same problems of complexity and lack of visualization that you get with a real animal or organ,” says Ingber.
Small is also better in the drug development process, during which researchers may screen hundreds of thousands of compounds to find a “hit” for a particular therapeutic need. The well plates used in drug screening, for example, may contain as many as 1,536 tiny wells on a playing-card-size piece of glass or plastic, allowing researchers to conduct that many individual experiments by dosing each cell with a different compound. A standard incubator system may include a hundred or more plates. To become practical components of drug development, organs on chips need to offer the ability to test similarly high numbers of potential drugs. But the larger the organ model, the fewer tests you can run; the more cells, drugs and reagents you need; and the more support hardware and technician time are required to keep the model alive.
Creating organ models on a microscale has been greatly facilitated by microfluidics, a technology developed in the 1990s that uses micropumps, valves and finely etched channels to manipulate the movement of fluids through a chip. Researchers are able to control the flow of blood, air, chemicals and drugs through the mini-organs to precisely mimic flow in the body. And a combination of microfluidic flow and other micro-architectural features of the devices enables the cells inside organ chips to live and thrive in three dimensions, not two. That’s important, says Joseph Vacanti, co-director of the Center for Regenerative Medicine at MGH, “because it’s the way cells normally exist in most parts of the body. Imagine an egg inside its shell compared to one that’s been cracked open and put in a frying pan. The egg in the shell feels at home in three-dimensional space. Cells are the same—they are most comfortable in three dimensions; they get completely different instructions for the behavior of their genes when they’re flat.”
ANOTHER ADVANTAGE OF ORGAN-ON-A-CHIP DEVICES is that they allow multiple cell and tissue types to function together in a way that replicates the dynamic interactions that take place in the body. “Tissues are groups of cells that exhibit specialized functions,” says Ingber. “Organs are more than one tissue coming together and forming new interfaces that have more complex functions than any single component on its own.”
In the human gut, for example, multiple layers of tissue combine to form a semipermeable barrier that allows useful nutrients in the alimentary canal to pass through and enter the bloodstream while deflecting potentially harmful bacteria and ingested toxins. The inner wall of the small intestine is lined with epithelial tissue covered in wrinkles and folds from which villi—microscopic fingerlike pieces of tissue—project, increasing the surface area for absorption. The villi are covered with absorptive enterocyte cells, along with goblet cells that produce mucus, which provide an additional layer of protection—and a potential barrier to the absorption of drugs taken orally. All of the cell types in the gut—enterocytes, goblet cells, enteroendocrine cells and Paneth cells—are derived from a type of stem cell that resides within the intestine.
The gut-on-a-chip developed by Ingber and colleagues at the Wyss Institute mimics this complex anatomy and physiology in ways that simple, static tissue cultures can’t. Inside the chip, two microfluidic channels are separated by a porous, flexible membrane. The membrane is coated with proteins that support 3-D cell growth and it’s seeded with human intestinal epithelial cells (from a cell line originally isolated from a colon cancer). When fluid is pumped slowly across both sides of the membrane, replicating the flow of liquid along the inside and outside of intestinal tissue in the body, the seeded cells not only differentiate into the four types found in the small intestine, but form tissue that spontaneously folds, develops villi, secretes mucus and even supports microbes commonly found in the small intestine. In a matter of days, a bunch of cells in a piece of plastic come to organize themselves into what is essentially a shrunken version of a small intestine.
Still another crucial part of creating a realistic organ proxy is the ability to reproduce a range of physical and other forces that influence normal cell development and behavior in the body. Cells in a human lung, for example, aren’t just lying undisturbed; they’re constantly expanding and contracting as you breathe. So the Wyss Institute lung-on-a-chip, about the size of a rubber eraser, mimics respiration with a vacuum pump system. In the gut-on-a-chip, a similar controller re-creates the wavelike peristaltic motion that moves food along the digestive tract.
A heart-on-a-chip being developed in the lab of Gordana Vunjak-Novakovic, professor of biomedical engineering at Columbia University, incorporates electrodes that act like mini pacemakers to “shock” the cells into beating, just as they would in a real heart. Electrical signals could also play an important role in modeling brain tissue, in which neurons communicate via electrical impulses, and for mimicking wound healing, which uses electrical signaling to guide cells to areas that need repair.
BUT ORGANS ON CHIPS AREN’T USEFUL ONLY as a way to create more realistic models of healthy, functioning organs; the miniature versions can also replicate what happens when things go wrong. A lung-on-a-chip created in the lab of Shuichi Takayama, professor of biomedical engineering and macromolecular science and engineering at the University of Michigan, for example, uses fluid mechanics to re-create the crackling stethoscope sounds associated with certain diseases of the lung. Takayama’s team was able to confirm that the sounds were associated with injury to the cells—suggesting that the model was accurately approximating what happens in a diseased lung—and to show how a therapeutic surfactant, which lowers the tension between liquids and solids, could reduce those injuries. “We could never do that in a dish culture,” says Takayama. “This is a dynamic mechanism that is hard to study in animal or human models.”
While these examples illustrate the complexity possible in organ-on-a-chip design, Takayama, Griffith and others are also interested in models that take a more stripped-down approach to imitating organs and their afflictions. “What’s interesting about this field is that there is a huge number of different problems, and lots of approaches are needed,” says Griffith. She is working, for example, on a model of liver function that fits hundreds of samples of 3-D liver tissue into the pits of a multiwell plate and uses micropumps to circulate nutrients and oxygen through the system.
Unlike “closed” systems, in which cells are enclosed in chips, Griffith’s open platform would allow researchers to work directly with the cultured tissue—for example, by chemically inducing inflammation and testing how different drugs interact with the inflamed tissue. That might help researchers quickly identify drugs that are likely to cause so-called idiosyncratic liver toxicity, for which preexisting liver inflammation may be a factor. “As engineers, we try to figure out what’s the easiest, lowest-cost, fastest way to get the information you need,” she says.
IN ANY KIND OF TISSUE ENGINEERING—whether the aim is to create transplantable organs or miniature organ-on-a-chip models—researchers sometimes depend on stem cells that can adapt themselves to the purpose at hand. But in the early days, scientists were limited in the types and quantities of cells they could use. They usually had to work with undifferentiated embryonic stem cells that were very hard to come by or tissue-specific adult stem cells that lacked needed flexibility. That changed with the discovery in 2006 of a genetic-engineering technique for “tricking” adult cells back into a pluripotent state. Such “induced pluripotent stem cells,” like other stem cells, are able to transform themselves into any type of cell in the body—making them ideal biological clay for tissue engineers to sculpt.
Yet because those cells also retain the DNA blueprint of their donor, it becomes at least theoretically possible to create a model not just of a heart—but of your heart. Or your tumor. “This opens the possibility of doing personalized disease models—personalized human chips,” says Nina Tandon, a researcher in Vunjak-Novakovic’s group at Columbia. With a grant from NIH, in collaboration with laboratories at MIT, Penn and Yale, the lab is developing an integrated heart-liver-vascular system for drug testing that will use stem cells derived from a patient’s skin cells, opening the possibility for personalized trials of potential treatments. If such systems became commonly available, you would no longer have to guess which of several available drugs would work best for you. You could simply test them all out on a micro-model of your heart and know for sure.
To gain widespread adoption in the academic and commercial research communities for personalized drug testing and other uses—and perhaps one day to replace millions of lab animals—organs on chips will need to be affordable and easy to use. They will also need to be qualified by the Food and Drug Administration as biomarkers—reliable tools for measuring biological processes or responses to drugs. Achieving that goal, says Ingber, will require proving that the devices are “as good as or better than animal models.”
That proof is beginning to accumulate. A 2012 paper co-authored by Ingber, for example, reported results from a study of a lung-on-a-chip model of pulmonary edema, which can affect cancer patients taking the drug interleukin-2. Ingber and his team found that a new class of drug being developed by GlaxoSmithKline could prevent pulmonary edema symptoms in the lung chip—and a separate study confirmed those results in animals. Developing the lung-on-a-chip took nearly four years, but it took less than two to modify it to demonstrate a human disease. “It gets faster as we go along because of what we learn in each prior system,” says Ingber.
ORGANS ON CHIPS EMERGED from efforts to engineer whole organs for replacement. But now, in turn, organ-on-a-chip research has helped advance tissue engineering’s original goal, with the closely controlled microenvironments of the chips yielding important insights into the forces that drive cells to become different types of tissue and assemble themselves into organs. And as newer versions of organs on chips get better at mimicking the function of real organs—a kidney’s ability to filter waste from the bloodstream, for example—the devices themselves may find their way into humans, replacing or augmenting underperforming organs.
“The ultimate goal,” says Vacanti, “is to put devices back into people, with the expectation that they will become part of a person and last as long as a person.” Such hybrid devices—part living, part nonliving—would have to be made of materials that wouldn’t be rejected by the body, and would need some kind of interface that enabled them to deal directly with blood flow. But Vacanti doesn’t think those are insurmountable hurdles. “It is feasible to extend the devices to help support functioning of the lungs, liver or kidney.”
Already, researchers at Vanderbilt University and the University of California, San Francisco, are collaborating on the construction of an implantable bio-artificial kidney that would contain microscopic filters and living kidney cells in a coffee-cup-size device that would work like a living dialysis machine inside the body—and provide most of the benefits of a kidney transplant. Increasingly, both strands of tissue engineering—creating organs ready for transplant, and organs on chips that may help alleviate the need for transplants—will continue to intertwine and inform each other. “I can’t believe how fast we’re moving,” says Ingber. “It’s very promising.”
Dossier
“Tissue Models: A Living System on a Chip,” Monya Baker, Nature, March 2011. This accessible review article provides a comprehensive overview of the history of organs on a chip and describes major directions in research. ?
“Engineering Challenges for Instrumenting and Controlling Integrated Organ-on-Chip Systems,” by John Wikswo et al., IEEE Transactions on Biomedical Engineering, March 2013. This technical article details multiple challenges involved in linking multiple organs on chips to create systems that realistically model human physiology.
“A Human Disease Model of Drug Toxicity–Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice,” Dongeun Huh et al., Science Translational Medicine, November 2012. A description of how researchers engineered a “sick” lung-on-a-chip that mimics the fluid swelling experienced by some patients taking the anticancer drug interleukin-2. Findings from this model led to the identification of potential drug therapies.
Stay on the frontiers of medicine