Published On May 3, 2014
FOR MORE THAN 10 YEARS, John Rinn, a cellular biologist who now teaches at Harvard Medical School, had been hunting for strands of genetic material that could explain the intricate orchestration of genes that switch on and off as embryos form and develop. But for just as many years, Rinn had heard that his quest was futile—that the objects he was seeking were bogus, mere phantoms that had no real functions.
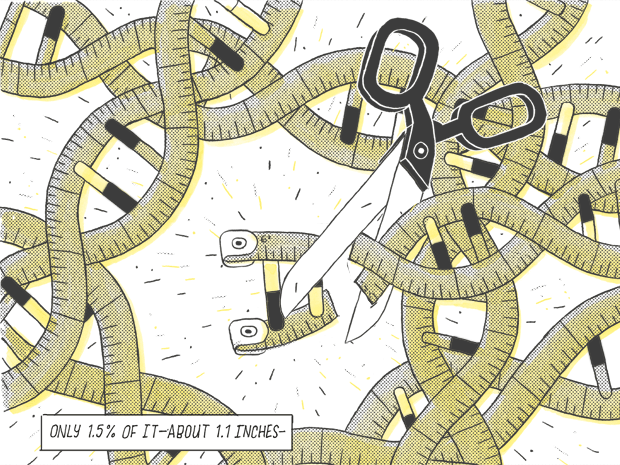
Rinn was at the forefront of a generation coming of age in the wake of the rather humbling draft publication of the human genome in 2000. It turns out that people have only about 20,000 genes—more than a fruit fly (14,000) but fewer than rice (51,000) and well short of the 100,000 or so that had been estimated for humans. Also, of the 3.5 billion base pairs of DNA in the human genome (pairs combining the chemicals designated as A, T, C and G), a mere 1.5% spell out genes that encode proteins—essential substances ranging from actin and myosin that build muscles to keratin for hair and fingernails.
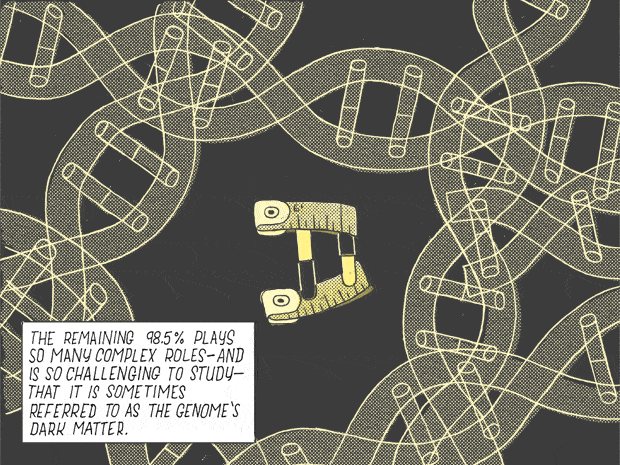
The remaining 98.5% of the genome is noncoding, meaning it does not produce proteins. Once dismissed as “junk DNA” and often dubbed “dark matter” in a nod to the characterization of the invisible matter that fills the universe, noncoding DNA is difficult to study and interpret. Much of it—just how much is a matter of hot debate—is transcribed into a complementary strand of RNA. But unlike the messenger RNA that carries information from genes to code proteins, this noncoding RNA (ncRNA) has no obvious purpose. However, Rinn and others believed that many long noncoding RNAs (lncRNAs) play crucial roles in the formation of embryos, development and disease.
The evidence they had, though, came mostly from studies of cells isolated in the laboratory—and uniform, cultured cells aren’t always reliable for predicting what happens in cells within the complex tissues and organs of a living animal. “I felt like I was at a poker table facing cowboys with guns,” says Rinn. “So I decided to put in all my chips and stake my career on the ultimate genetic test. If I lost, it would answer an important question: Yes, this noncoding RNA is junk after all. But if the cards came up right…”
Rinn and his colleagues’ gamble was to select 18 lncRNAs that prior research suggested might have biological importance, “knock out” each one in breeding a separate line of mice, and observe the impact on the animals for a study in the Dec. 31, 2013 eLife journal. Mice born without these noncoding RNAs had a variety of defects in anatomy, development and viability. Some had stunted growth, and quite a few died before birth. One had kidney and lung defects, another had epilepsy and seizures, and another was missing part of its brain. “We still don’t know what these lncRNAs are doing or how they work,” Rinn says. “We just know that if we take them away, bad things happen.”
That’s important, because a major impetus for exploring the human genome has been to identify genes involved in human disorders—with the ultimate goal of finding new treatments. Massive genome-wide association studies (GWAS) have compared the genetic makeup of healthy people with the genes of those who have heart disease, diabetes, obesity, Alzheimer’s, arthritis and other conditions. Intriguingly, though, many GWAS pointed toward noncoding areas in which previously undetected regulatory elements might lie. To help determine what’s out there, the National Human Genome Research Institute has funded the international Encyclopedia of DNA Elements (ENCODE) Consortium.
In this developing picture, the noncoding genome appears to yield both subtle and profound effects, from helping faces and hands emerge with human characteristics to causing cancer or physical defects. While many findings have their detractors, the cumulative effect is forcing scientists to rethink some of their most basic assumptions about genetics.
The first solid evidence that a long noncoding RNA might be important involved the X chromosome in females. Unlike men, who inherit both an X and a Y chromosome, females have two Xs, and one must be inactivated to avoid problems that would arise if both chromosomes expressed all of their genes. That silencing happens in embryos through a tricky procedure involving something called Xist, once thought to be a protein but in fact a lncRNA.
In 1991, researchers discovered that Xist “coats” the X chromosome that’s designated to be silent, shielding it from the factors that would activate its genes. “But it took a very long time to prove that Xist works as a long noncoding RNA,” says Jeannie Lee, a molecular biologist at Massachusetts General Hospital who observed the first hint of this idea in 2006. The scenario that she and others have worked out is that, early in the development of an embryo, the two X chromosomes temporarily align, making physical contact that allows communication between an area on each chromosome known as the “X inactivation center” (Xic) to determine which of the two to inactivate. Then the Xist RNA unfurls from the Xic of the designated silent chromosome and binds to a protein (polycomb repressive complex 2 or PRC2) that suppresses the activity of many of that chromosome’s genes. The silent X condenses, becoming a tightly packed shape called a Barr body.
Yet when a cell divides and must duplicate its chromosomes, the Xist/PRC2 shroud falls off and must be reassembled in each daughter cell. But that’s a faster, easier process than it was in the early embryo. Each new cell retains a memory of which X is to be silenced, and the Xist cloud settles evenly over the entire chromosome.
AT FIRST THE REMARKABLE ROLE of Xist as a lncRNA was considered an exception, but soon more lncRNAs were suspected to have crucial functions. Despite the debate about lncRNAs’ significance, it was generally agreed that all noncoding regulatory elements acted on the same chromosomes on which the RNA’s own sequences were located: They acted in cis, from Latin for “the same side.”

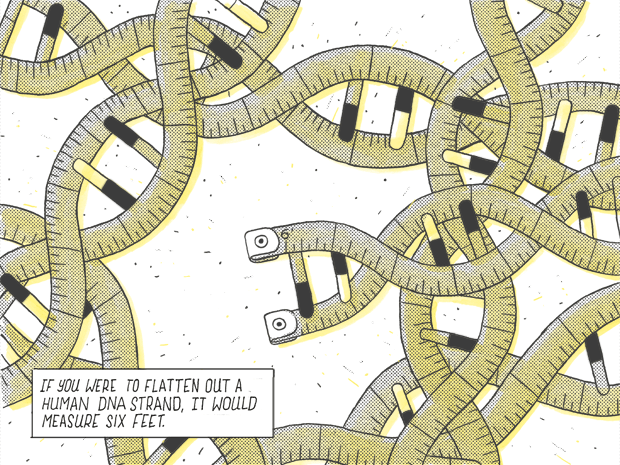
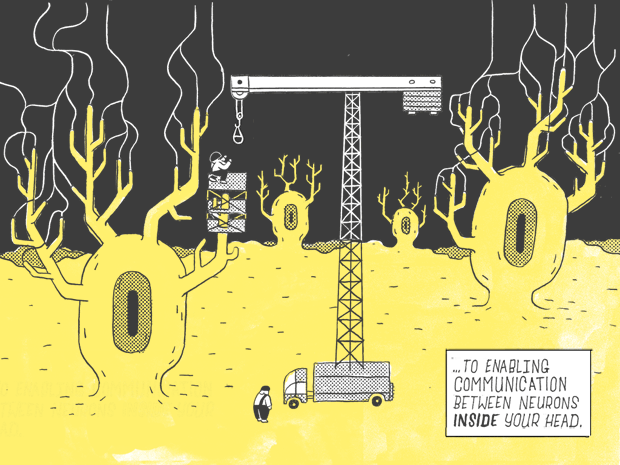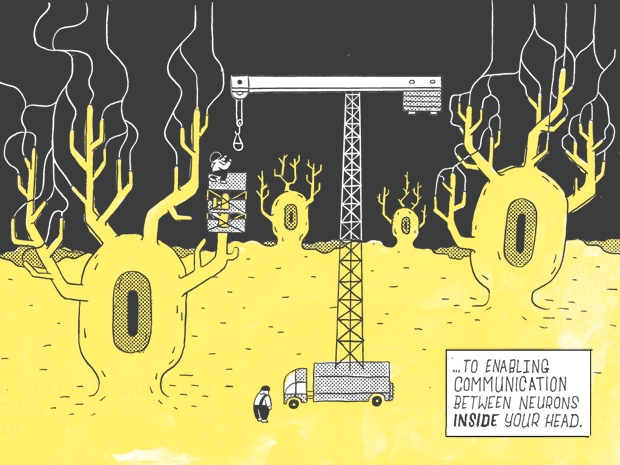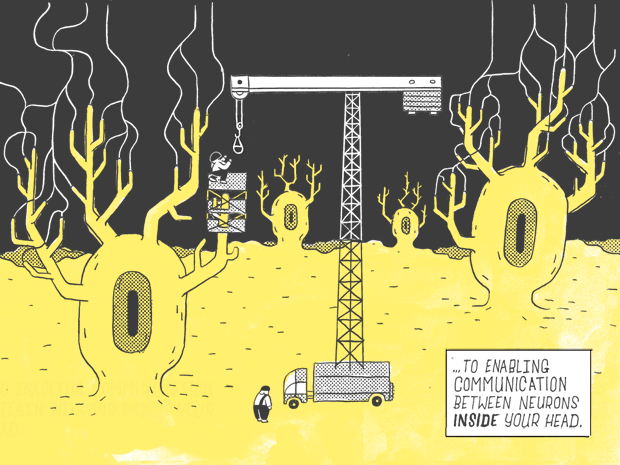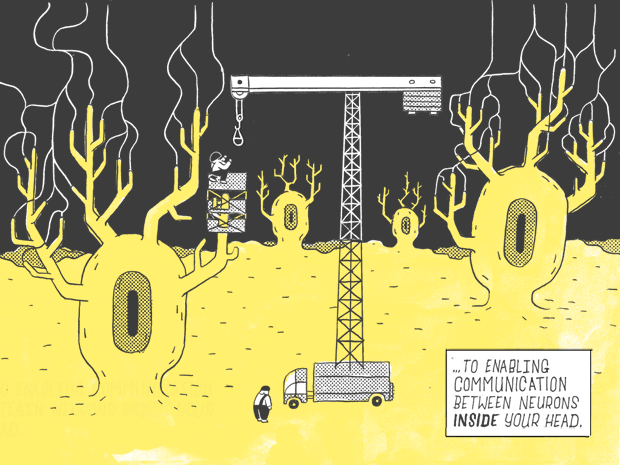


If you were to flatten out a human DNA strand, it would measure six feet.
Only 1.5% of it — about 1.1 inches.
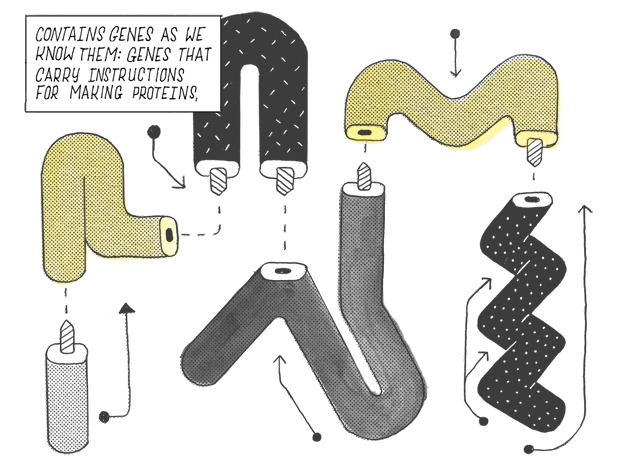
Contains genes as we know them: genes that carry instructions for making proteins.
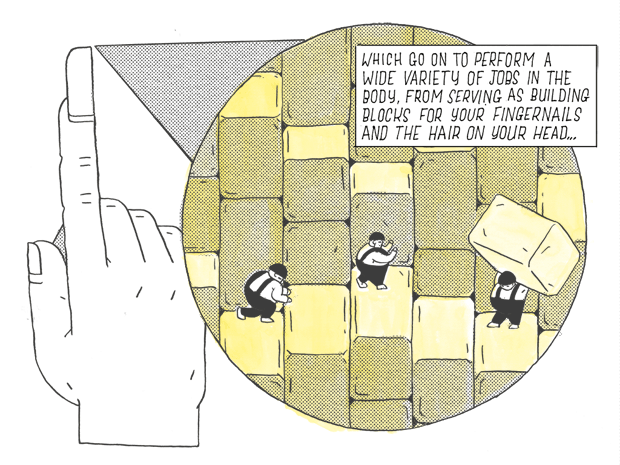
Which go on to perform a wide variety of jobs in the body, from serving as building blocks, for fingernails and the hair on your head…
…to enabling communication with the neurons inside your head.
The remaining 98.5% plays so many complex roles — and is so challenging to study — that it is sometimes refer to as the genome’s dark matter.
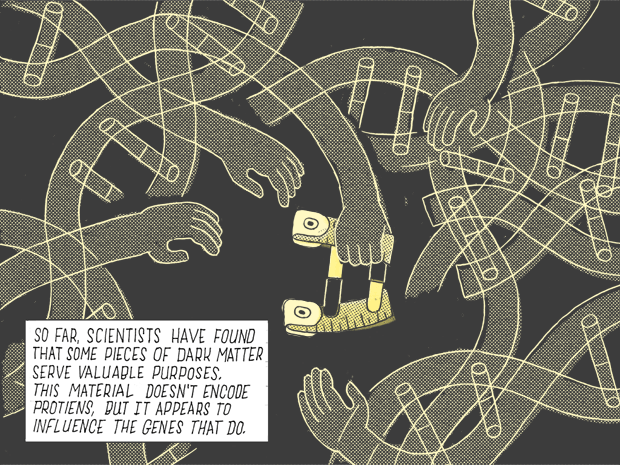
So far, scientists have found that some pieces of dark matter serve valuable purposes. This matter doesn’t encode proteins, but it appears to influence the genes that do.
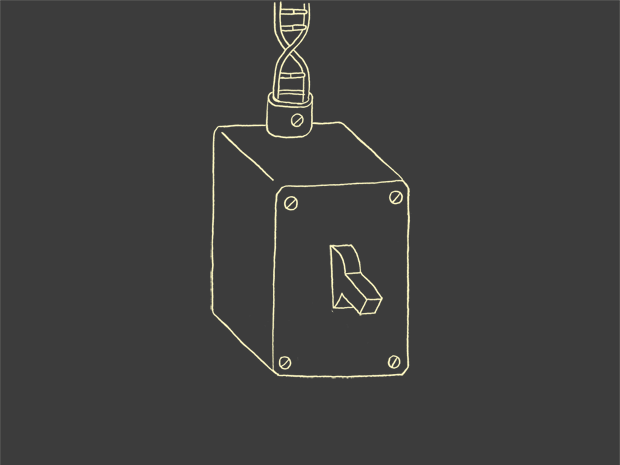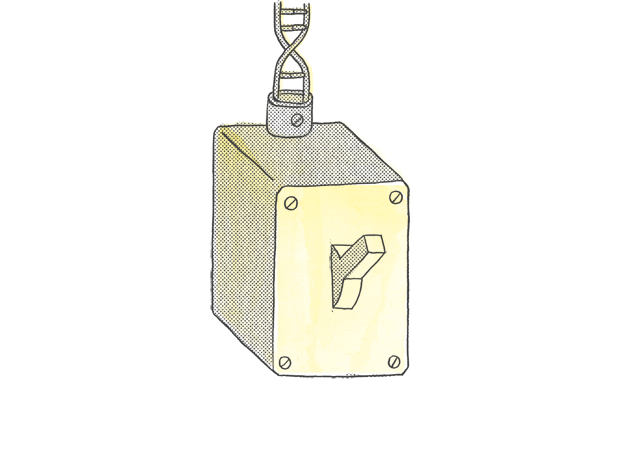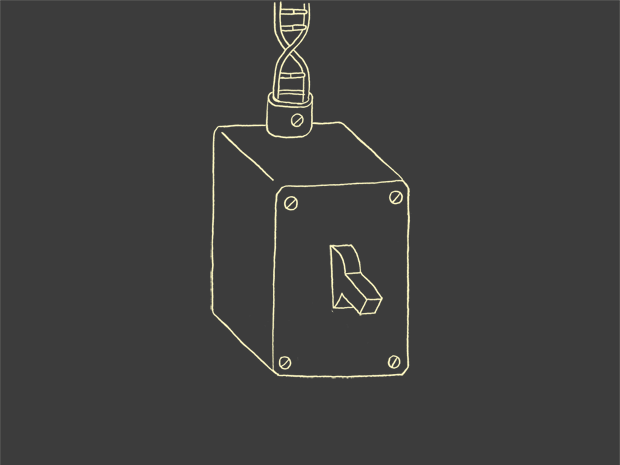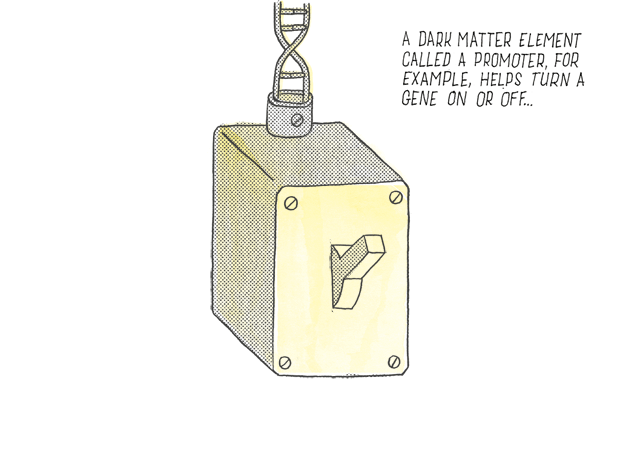
A dark element called a promoter for example, helps turn a gene on or off …
…Whereas another element called an enhancer acts as a dimmer switch on a gene.
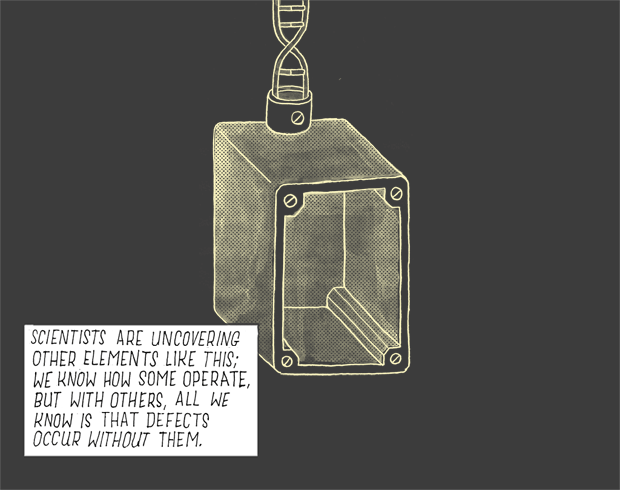
Scientists are uncovering other elements like this; we know how some operate, but with others, all we know is that defects occur without them.
We are only just beginning to figure out whether a tiny percentage of these elements have any real function — or if most do.
In 2007, however, Rinn, then a postdoctoral fellow under Howard Chang at Stanford University School of Medicine, and his colleagues were astonished to discover a lncRNA that worked in trans—on a different chromosome. They had identified a lncRNA called HOTAIR in developmentally important HOX gene clusters. HOX clusters, from A to D, contain genes that define the body plan from head to toe, as reflected in the progressively detailed features of a growing fetus. HOTAIR is located near the HOXC cluster on chromosome 12, so Rinn thought that by deleting its DNA he could see whether one of the nearby HOXC genes would be affected. The deletion did have an impact, but on HOXD13 on chromosome 2.
HERE, too, what first seemed like an exception—to the rule of in cis regulatory elements—now appears more common, and in trying to determine how such elements might wield their influence from afar, researchers have also had to rethink other basic assumptions. For example, they had typically thought of the genome as a linear sequence of DNA bases (A, T, C and G) that align in a long chain, with various sections spelling out instructional recipes for proteins. But the reality turns out to be much more complex.
“You can think of the genome as having three layers of information,” says Job Dekker, a biochemist and molecular pharmacologist at the University of Massachusetts Medical School. The first layer involves the familiar linear DNA sequence. But if all of the DNA in the genome sequence were straightened out, it would extend to six feet. How does that long strand get squeezed inside a cell nucleus that’s a mere 10 to 20 microns (millionths of a meter) in diameter without becoming hopelessly entangled? How do the various factors that switch on a gene find their target? “That’s the biggest challenge for cells,” Dekker says, “and for a very long time we weren’t able to figure out how they solve it.”
The solution lies in how the DNA is tightly wound around spools called histones, which are packaged with a coating of chemicals called chromatin. Chromatin is decorated with “flags” indicating where genes, lncRNAs and other regulatory elements are tucked away inside—and that system comprises the second layer of genomic information. It enables other molecules to locate those elements, causing the chromatin to open or close depending on whether an element needs to be activated or silenced.
This layer of information is tied to the third layer, which concerns how the chromosomes themselves fold and loop within the cell nucleus. That looping can bring genes and noncoding regulatory elements that lie far apart in the linear sequence (and even on different chromosomes) into close proximity and even physical contact. That three-dimensional physical interaction (the third layer), in turn, may change the chromatin configuration (affecting the second layer) and thus influence the activity of genes (written in the first, linear layer of information).
One way the three layers interact involves the relationships between two noncoding elements, promoters and enhancers, and the genes they influence. “The more we learn about regulatory elements, the more we realize the categories we have for them are more fluid than we thought,” says James Noonan, a geneticist at Yale University. Nevertheless, it’s generally understood that a promoter acts like a light switch that is activated by molecules called transcription factors to turn on a gene. Enhancers act as dimmer dials; they recruit transcription factors and guide them to a targeted promoter, and that interaction modulates gene expression. However, enhancers often lie at an immense distance from the promoters they target. They come together thanks to the looping of DNA strands, but exactly how that works remains a mystery.
Each enhancer may bind many transcription factors, and each gene may be regulated by many enhancers—and every cell type has different sets of transcription factors and enhancers. “Enhancers are highly specific for certain tissues, cell types, time points in development, and environmental conditions or external stimuli,” says Axel Visel, a staff scientist in the genomics division at Lawrence Berkeley National Laboratory in Berkeley, Calif.
Visel notes that enhancers, because they have been conserved through the evolution of species, are thought to have played a fundamental role in those gradual genetic changes. “Nature held on to these elements because they do something important,” says Visel, who is currently studying evolutionarily conserved enhancers that affect the formation of the face, brain, heart and other organ systems. Meanwhile, though, some enhancers were changed, and others were occasionally added in separating humans from their ancestral species. Yale’s Noonan thinks such “human gain” enhancers may control some of people’s most distinguishing features: hands and feet, dexterous fingers, upright posture, face, cranium and brain. “To understand what makes humans distinct from other species, we need to understand how human development is different,” says Noonan.
In his work, Noonan focused first on limb development because it was comparatively well understood and because it’s an area in which humans evolved quite differently than other species. Ina study published in Cell in July 2013, he and his colleagues compared embryonic tissue in humans, rhesus monkeys and mice during four developmental stages. He found several thousand enhancers that were more active in humans, including many that were associated with genes involved in the formation and shapes of tendons, cartilage, the big toe and height. Now he’s testing whether the human versions of these enhancers will affect development in mouse models. Next he plans to apply this method to the development of the brain.
ACCUMULATING RESEARCH SUGGESTS that the role of the noncoding genome might be even more important than protein-coding genes in understanding how human diseases develop. For example, in February 2013, two groups independently identified the same two closely related mutations in a promoter that regulates telomerase, an enzyme that almost all cancers exploit so that they can divide indefinitely. More than 70% of melanoma tumors had this promoter mutation. That kickstarted a series of additional studies that have found the same two promoter mutations in many cancers—in 44% of hepatocellular carcinomas, 66% of bladder cancers and 83% of glioblastomas. In all cases, those mutations are more prevalent than any gene mutation that has been identified.
Enhancers are also being linked to disorders in which mutations in protein-coding genes have proven hard to find. For example, a genome-wide association study found a sequence that significantly increases the risk of being born with a cleft of the lip or palate. “But there was not a single protein-coding gene in that sequence,” says Visel. In a 2013 Science study, however, he reported that he found in this region at least two enhancers that are implicated in facial development and may act on genes that sit outside the region. It may be that the gene itself isn’t mutated in the disease but rather that the problem lies with an enhancer that regulates the gene.
Similarly, Lee of MGH has linked the lncRNA Xist to blood cancer in mice. Could those cancers have defective X inactivation? In a February 2013 article in Cell, she described deleting Xist in embryonic blood stem cells in mice, causing all of the blood cells in the developing female mice to have two active X chromosomes. The upshot? All of the female mice missing Xist developed blood cancers. But none of the male mice without Xist had problems—because, having only one X chromosome, they don’t need X inactivation. “If we could reintroduce Xist or use some other technique to inactivate the second X chromosome, it might suppress these cancers,” Lee says.
Meanwhile, researchers at University of Massachusetts Medical School recently exploited Xist’s capacity to silence an entire chromosome for a study of Down syndrome, in which children are born with an extra copy of chromosome 21. Working with cells in culture, the researchers inserted Xist and saw it create the “cloud” that silences the X chromosome, and the same condensed form that kept its genes shut down.
IT REMAINS TO BE SEEN how Rinn’s bet on the importance of lncRNAs will play out. So far, he and others have studied only a relative handful of what could be thousands of these noncoding RNAs, and scientists are also only beginning to explore other regulatory elements. Even if most of the known elements have a significant impact on human biology, together they account for but a tiny part of the genome. Still, the importance of known noncoding regulatory elements may rival that of the 1.5% of the genome that contains protein-coding genes.
What’s important for now is that many noncoding elements merit exploring in animal models of human diseases. The results of those tests, many hope, will provide clearer glimpses into our vast genome and bring us closer to the secrets we had hoped to discover when we began translating the human “book of life” in 2000. So far those glimpses inspire the same sense of wonder as the Fermi Gamma-ray Space Telescope that was sent out to explore what the Fermilab calls the “numerous exotic and beautiful phenomena” in the dark matter of the material universe.
Dossier
“Genome Regulation by Long Noncoding RNAs,” by John L. Rinn and Howard Y. Chang, Annual Review of Biochemistry, July 2012. The authors, who have made major contributions to the understanding of long noncoding RNAs, review technologies enabling these studies and highlight emerging themes.
“The Evolution of Lineage-Specific Regulatory Activities in the Human Embryonic Limb,” by James P. Noonan et al., Cell, July 3, 2013. Noonan describes enhancers associated with the embryonic development of the distinctive human hand and foot.
“Xist RNA Is a Potent Suppressor of Hematologic Cancer in Mice,” by Jeannie T. Lee et al., Cell, Feb. 14, 2013. Some breast cancers have extra copies of the X chromosome, suggesting that these cancers may have malfunctioning Xist, a long noncoding RNA responsible for silencing the second X chromosome in females. Here, Lee and colleagues investigate whether Xist dysregulation might cause cancer, using blood cancer in mice as a test case.
Stay on the frontiers of medicine
Related Stories
- Wanted: A Better Analogy
Why geneticists hate the term dark matter.