Published On January 15, 2009
THE LECTURE WAS ABOUT JELLYFISH, but Martin Chalfie was daydreaming about worms. After the speaker described a protein—green fluorescent protein, or GFP—that makes jellyfish glow in the dark, “I got very excited and didn’t listen to another word,” Chalfie remembers. It was 1988, and Chalfie, a biological science professor at Columbia University, found himself thinking about the recently discovered protein: “What a wonderful compound to put into C. elegans.”
At the time, the nematode Caenorhabditis elegans, or C. elegans, had attracted a small, fervent circle of researchers who had come to recognize the worm as a simple model for some of the most complex processes at work in human cells. Transparent and a mere 1 millimeter long, the worm was thought to share many fundamental genetic characteristics with people, though scientists estimated that the species’ evolutionary paths had diverged some 800 million years earlier. But the techniques to explore those similarities had barely been born.
His mind wandering at the lecture, Chalfie envisioned the glowing protein, somehow inserted into the worm and linked to other proteins, serving as a kind of green highlighter marking the tiniest of cellular processes. If he could figure out a way to insert GFP without needing any other compound from the jellyfish, it would be a breakthrough for studying C. elegans and its nearly 1,000 cells, and it would add to the worm’s renown as an ideal model organism for unraveling the mysteries of human genetics.
Chalfie’s hunch proved prescient, though it took several years of painstaking effort to get GFP working in the worm. And his achievement with C. elegans was just one of many. During the past half-century, the nearly microscopic, see-through worm has been so closely observed that scientists have a nearly exhaustive knowledge of its brain, nervous system, digestive tract, musculature and reproductive system. That knowledge, in turn, has greatly expanded the understanding of human systems and processes.
All this groundbreaking worm work has earned its champions three Nobel Prizes during the past six years (including one, in 2008, for Chalfie, who shared the chemistry prize for pioneering the use of GFP), with more likely to come. “C. elegans has led to fundamental changes in our understanding of life,” says Chalfie, who continues to use the worm to study nerve cell development and function. Meanwhile, the community of worm researchers, once almost as diminutive as its subject, has expanded exponentially, with scientists now using C. elegans to gain insight into the mechanisms of aging, Alzheimer’s disease, stroke, cancer, retinitis, diabetes, kidney disease and other disorders.
“The worm poses questions we could never have even thought about without it,” says Robert Waterston, a professor of genome science at the University of Washington’s School of Medicine in Seattle. And the answers keep coming, as C. elegans guides researchers into a previously unimagined universe of regulatory molecules that could revolutionize medicine and drug development.
FOR MILLENNIA, C. ELEGANS LIVED MOSTLY UNNOTICED in its natural habitats of soil and water or as a parasite in snails and other animals and plants found across Europe, North America and Australia. Then, in 1954, in Bristol, England, a biologist named Warwick Nicholas plucked it from obscurity. Nicholas, one of several biologists who had been considering nematodes as a possible model organism, was given a colony of C. elegans (first classified in 1900 by French biologist Emile Maupas) that had been thriving in a tray of mushroom compost. He succeeded in growing cultures of C. elegans, which he took with him in sealed test tubes to California when he moved there. One of several nematode strains Nicholas was examining, C. elegansproved particularly useful. And though Nicholas’s work with the worm, which focused on its development and reproduction, was only a prelude, the colony he established became the progenitor of almost every one of the trillions of C. elegans that have been making themselves at home in laboratories around the world ever since.
In 1964 descendants of the Bristol generation ended up back in England, in the laboratory of Sydney Brenner, an investigator at the Medical Research Council in Cambridge, who had already made groundbreaking contributions to the emerging field of molecular biology. Brenner had discovered basic genetic structures in simpler organisms (mostly bacteria and viruses) and now he wanted to find out how a higher organism used the instructions encoded in DNA to build its three-dimensional self. He decided he needed an animal model to tackle these complex biological problems, and after trying out more than 60 nematode species, he settled on the strain of C. elegans he’d obtained from the University of California at Berkeley.
Because C. elegans is transparent, it can be observed through a microscope as it grows from an egg, with its cells dividing repeatedly to form the worm’s organs, nerves and muscles. And because the worm has a life span of just two to three weeks and can produce 100,000 progeny in 10 days, experiments could be done quickly and, just as important, in a space as small as Brenner’s cramped Cambridge laboratory. To do similar work with mice, he would have needed a building several stories high.
Brenner’s first goal was to discover how the worm’s behavior is wired into its nervous system. That meant taking the organism apart physically and charting all the connections and characteristics that make the worm a worm, a project that seemed quixotic to many of Brenner’s colleagues.
“They thought it was a blind alley,” says David Hall, a professor of neuroscience at Albert Einstein College of Medicine in New York City and the editor of WormAtlas, an Internet database devoted to C. elegans research. But Brenner persisted, and though the quest took 10 years, he and his research group succeeded in producing the first map of the worm’s anatomy. They detailed the development of every cell, starting with the egg and progressing to the 959 body cells of the adult worm. They also created a “wiring” diagram of every neuron in C. elegans’s 302-cell nervous system.
It was a hugely laborious process that involved slicing the worm into sections thin enough to fit under the lenses of an electron microscope. Researchers took more than 30,000 photographs to identify and trace the visible structures. Then they pieced together the images to form somewhat ill-fitting cross sections of C. elegans’s skin, nervous system and other organs. Notes had to be taken by hand and were bound into books. “It’s hard to believe how primitive things were,” says Hall, who keeps many of the early photographs and notebooks in his laboratory.
This map of C. elegans’s cells and nervous system gave researchers a tool for testing theories about how the organism grows and behaves and for identifying the genes involved by comparing the makeup of normal worms with that of genetically manipulated strains. Experiment by experiment, Brenner learned more and more about the worm and its genetic makeup—for example, that there are about 100 mutations that can cause problems with wriggling. Some of those mutations lead to mistakes in the way the nervous system grows, whereas others keep the system from functioning properly after it is formed.
Getting to know this single species so intimately led to insights into key biological processes. For one thing, scientists noticed that during the course of worm development, 131 cells self-destruct. Robert Horvitz, who came to Brenner’s laboratory as a postdoctoral fellow in 1974, returned to the Massachusetts Institute of Technology four years later and continued to study this phenomenon. Horvitz went on to prove that cell death, or apoptosis, is actually programmed into some of the worm’s genes as a fundamental part of its development. And most of those genes turn out to have counterparts in humans.
Programmed cell death explains why people end up with fingers and toes but without the webs that exist in fetuses. And it provides clues about the biological processes that lead to cancer, which may result from abnormalities in cells that stave off apoptosis, making themselves immortal. (Horvitz, Brenner and John Sulston, a British biologist who now chairs the Institute for Science, Ethics and Innovation at the University of Manchester, shared a 2002 Nobel for this work.)
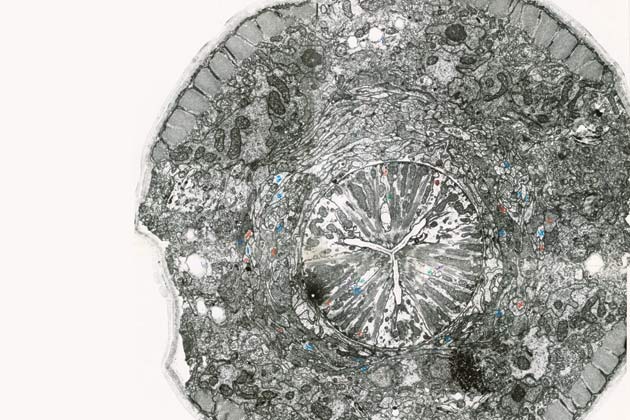
Mapping the more or less visible parts of C. elegans was an enormous achievement. But to exploit the full potential of the worm’s unlikely likeness to humans, preserved through eons of evolution, scientists needed to understand the species at an even more fundamental level. Brenner’s vision was to go deeper and catalogue the genetic structures inside every cell, to find out not only how single genes work but also how they function in combination. The way the chemical building blocks of DNA—adenine, thymine, guanine and cytosine, or A, T, G and C—are paired and sequenced determines the proteins a cell makes, so unraveling the entire genome for C. elegans would essentially spell out the instructions for its life.
Beginning in the early 1980s, Waterston, then at Washington University’s School of Medicine in St. Louis, and Sulston worked with a team of researchers to break C. elegans’s DNA into 17,000 overlapping fragments to create a map. To do so, they had to put the fragments back in order, “a little like knowing chapters in a book,” Waterston says, with a process called gel electrophoresis. After the fragments were treated with enzymes and radioactive compounds, they were then separated and the results were captured on X-ray film. The resulting images showed dark bands of DNA as if on a bar code. The researchers used computers to match the images and put the fragments in order. Waterston and Sulston then determined the entire genome sequence.
“Before the map, it could take someone years to find just one gene,” says Waterston; afterward, the task could be done in a matter of weeks. Like the anatomical blueprint of the worm, this was a first, providing a panoramic genetic view of an animal, with a detailed reconstruction of its nearly 20,000 genes, 6 chromosomes and some 100 million base pairs of DNA. The completed worm sequence triggered a revolution in science and medicine.
Waterston and Sulston also led the way in compiling the human genome, which was finished five years later, and today researchers are detailing the genomes of chimpanzees, mice and a host of other organisms. What’s more, the kind of gene sequencing Waterston and his colleagues pioneered is now being done by biotechnology companies and academic research centers with equipment that can rapidly interpret thousands of genetic sequences. As a result, the field of comparative genomics, which compares the genetic structures of different species and strains, has opened untold areas of research, ranging from studying evolutionary changes among organisms to developing strategies for combating human disease.
HOMO SAPIENS AND CAENORHABDITIS ELEGANS ARE NOT particularly close genetic cousins. Fewer than half of the worm’s genes have human counterparts, far less than, say, chimpanzees, which have about 95% of the same genetic material as humans. Still, exploiting those similarities continues to prove extraordinarily fruitful. In the latest, potentially game-changing worm research, scientists such as Gary Ruvkun, a molecular biologist at the Massachusetts General Hospital and a professor of genetics at Harvard Medical School, are focusing on genetic material known as RNA, which has many flexible forms that take on wide-ranging regulatory roles inside cells. Understanding and manipulating those functions could lead to new ways to treat disease.
Very small pieces of this material, called microRNA, have proven particularly interesting; the first hints of their importance came in the early 1990s, when Ruvkun and Victor Ambros, now a professor of molecular medicine at the University of Massachusetts Medical School, were studying how C. elegans develops into its adult form from newly hatched larvae. Research on flies had shown that particular genes instruct embryos where to grow wings, legs and other body parts. Suspecting that comparable genes in the worm specified the timing of similar developmental events, Ambros focused on lin-4, a worm gene Brenner had discovered that allows immature worms to advance past a particular developmental stage. Researchers already knew that animals with a defective version oflin-4 can’t become adults. Ambros found that worms with inactive lin-4 get stuck repeating early larval stages, and those lacking a different gene, lin-14, skip ahead, with cells dividing in patterns more characteristic of later stages of development. In a series of experiments, he and Ruvkun showed how the genes collaborate—that at an appropriate time lin-4 blocks lin-14 activity, allowing worms to continue normal development, and that very small strands of RNA play a role in that process.
In 1998, Andrew Fire, a biologist and professor of pathology and genetics at the Stanford University School of Medicine who had also worked with Brenner, and Craig Mello, a professor of molecular medicine at the University of Massachusetts Medical School, made the next leap forward, unveiling work they’d done in C. elegans that explained the role of RNA interference, or RNAi. RNA ordinarily transfers genetic instructions from DNA in a cell’s nucleus to the surrounding cytoplasm, where those codes guide the construction of proteins. Fire and Mello injected the worm with gene-carrying fragments of RNA that disrupted this normal process, preventing the synthesis of the protein associated with the gene on the fragment (work for which they won a Nobel in 2006). This RNA interference, which occurs naturally in C. elegans and other organisms, including humans, can be manipulated for research purposes, allowing scientists to silence one gene at a time with great precision.
Ruvkun’s laboratory has used RNAi to work systematically through the C. elegans genome, inactivating individual genes and observing the impact on the worm. Some of this work has focused on worm metabolism, and among the almost 17,000 worm genes his team has tested, 305 inactivations caused a decrease in the worm’s body fat, while 112 resulted in increased fat storage. About half of those 400-plus fat genes have human counterparts, and if those too could be manipulated, it might provide an avenue for developing medications to treat obesity and related diseases such as diabetes. “If worm obesity were a health problem, we‘d have a cure,” quips Ruvkun, who says this work has helped narrow the pool of genes that might be involved in regulating human fat storage to a virtual droplet: just a few dozen genes (out of some 20,000 in the human genome).
Fire and Mello’s discovery also shed light on the part played by microRNA. In C. elegans, microRNA adjusts the production of proteins that are needed at one stage of a cell’s life but must be absent during others. “Perhaps RNAi and microRNA are more similar than we currently appreciate,” says Ruvkun, who in 2008 shared the Albert Lasker Basic Medical Research Award (sometimes called the American Nobel) with Ambros and the University of Cambridge’s David Baulcombe for their work in C. elegans. “What we know now is the tip of the iceberg.”
These RNA mechanisms could lead to a completely new, finely tuned form of disease therapy, Ruvkun says. Whereas current drugs act on proteins, RNA therapy could come in a step earlier, acting on the expression of DNA itself before a problem protein is produced, potentially preventing or reversing diseases. According to Ruvkun, recent studies suggest the human genome contains more than 500 and perhaps as many as 1,000 microRNAs that, collectively, might control a third of all protein-producing genes. (Not all genes produce proteins; a significant fraction play roles in determining embryonic development, blood-cell specialization and a wide range of other physiological functions.)
Researchers are now trying to develop drugs that work by selectively blocking microRNAs. A number of labs have been working to compare microRNAs produced in cancerous tumors with those in normal tissue. Ideas developed in C. elegans can then be tested in higher organisms such as mice, whose response to a drug or other treatment may be more directly relevant to people—perhaps leading, Ruvkun speculates, to future generations of drugs, particularly for cancer treatment, that involve microRNAs. “This is the hottest hunch right now, not just among worm researchers but in the larger scientific community, and there are probably 1,000 people working on it,” Ruvkun says. “Once again, the worm has put us on the right track.”
Dossier
In the Beginning Was the Worm, by Andrew Brown (Columbia University Press, 2003). Brown, a science journalist, tells the compelling story of Sydney Brenner’s worm project and of the vision, personalities and work of the first generation of worm researchers.
“The Perfect Storm of Tiny RNAs,” by Gary Ruvkun, Nature Medicine, October 2008. The 2008 Lasker Award winner details how C. elegans research led to the breakthrough discovery of a universe of tiny regulatory RNAs that were initially considered “worm curiosities” until they were also discovered in plants, and why this worm is important to the future of medicine.
wormatlas.org and wormbase.org. WormAtlas describes every cell and tissue of C. elegans and includes information developed for teaching purposes. WormBase houses information about all C. elegans genes and details research in such categories as cell biology, neurobiology, evolution and disease models.
Stay on the frontiers of medicine